Solving the structure of a multi-enzyme bacterial complex may help antibiotics do their job against the deadly disease
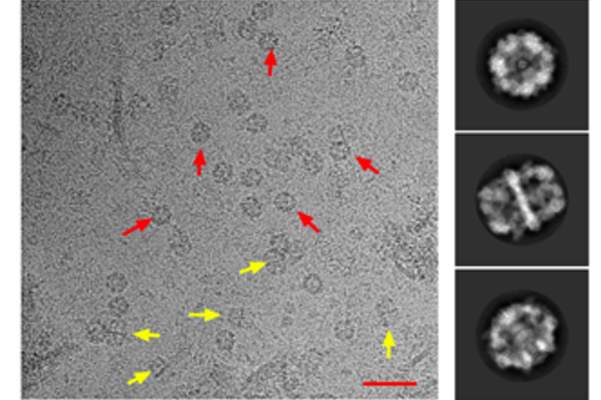
Cryo-EM micrograph of Mtb FAS-I particles. FAS-I particles are clearly visible in the micrographs. Scale bar represents 50 nm.
Tuberculosis (TB) killed an estimated 1.7 million people and newly infected 10 million in 2016 alone, according to World Health Organization figures. As some strains are becoming resistant to all known antibiotics, new approaches are desperately needed. One potential target for drug design is the thick, waxy outer coating that conceals Mycobacteria tuberculosis from the immune system and keeps antibiotics from penetrating the cell. A collaborative effort at the Weizmann Institute of Science recently employed a cutting-edge electron microscopy method to reveal, in unprecedented resolution, the structure of a large complex for synthesizing the building blocks of this coating. Their findings, published in Nature Communications, may point the way toward fighting antibiotic resistance.
Dr. Ron Diskin of the Institute’s Structural Biology Department explains that most living cells have one of two systems called fatty acid synthases (FAS), which are ensembles comprised of several enzymes. These systems produce the fat-based molecules that make up the cell membranes. Animals and fungi tend to have type 1 (FAS-I), bacteria and archaea type 2 (FAS-II). But the TB bacterium has both FAS-I and FAS-II, and these two cooperate to produce fatty acids that are extra-long — up to 90 carbon links in the chain, in contrast to the 16-carbon fatty acids in our cells. Prof. Oren Zimhony of the Kaplan Medical Center, who is affiliated with the School of Medicine of the Hebrew University of Jerusalem, had amassed evidence over a decade of research that the Mycobacteriatuberculosis FAS-I should be an excellent drug target. Such drugs can be designed with the aid of computer algorithms, but first one needs to know exactly how the complexes of enzyme molecules are configured and where their weak points lie. It is the large size of the FAS-I complexes that has made them especially challenging to map out and study.
First challenge: FAS purification
Zimhony’s first challenge was to purify the Mycobacterial FAS-I system so that it could undergo structural analysis. Several years ago, Zimhony approached Profs. Zippora Shakked and Yigal Burstein of the Weizmann Institute’s Structural Biology and Organic Chemistry Departments, respectively, and the two agreed to support this highly challenging research. Zimhony started experiments with a talented biochemist, the late Alon Schwartz, as well as with Dr. Moshe Peretz of the Structural Biology Department. In the resulting PLoS One paper, Zimhony summarized the method developed by this team for purifying FAS-I from mycobacteria, and they presented experimental validation for FAS-I activation. But despite their efforts, they did not succeed in obtaining high-quality FAS-I crystals for structural analysis by X-ray crystallography. Preliminary electron microscopy (EM) analysis, however, indicated a tantalizing potential for this technique to get them all the way to structurally analyzing FAS-I.

(l-r), front: Jacob Grunwald, Dr. Szilvia Baron, Dr. Ron Diskin, Prof. Oren Zimhony and Dr. Nadav Elad. back: Dr. Shira Albeck, Gal Raviv, Dr. Yoav Peleg and Prof. Zippora Shakked
Zimhony and Shakked then turned to Diskin, and together they decided to use an approach known as ‘single particle cryo-EM” to attempt the challenge of solving the structure of this enzyme ensemble. Together with Dr. Nadav Elad of Chemical Research Support and others, fine carbon grids containing FAS-I molecules in vitrified ice were prepared and then transported to a special electron microscope in Heidelberg, Germany, where Elad oversaw the imaging. The team then returned to the computer screen and algorithms to combine the Heidelberg data into high-resolution, three-dimensional images revealing the FAS-I structure at near-atomic resolution. The researchers compared the structure they had obtained with that of another known FAS-I system, in fungi, and discovered that there were more differences between the two than they had expected, especially in the parts in which the fatty acid chains are manufactured. They noted deeper clefts and alterations in the charges facing the active sites, which they think help make the Mycobacterial FAS-I system sturdy enough to produce the longer chains.
Further challenges: drug design and testing
“This is exactly the level of resolution at which we can begin to design drugs,” says Diskin, “as we can see not just the general shape, but really where and how a drug molecule needs to bind in order to inhibit an enzyme’s activity.
“At least one version of such a drug might already exist,” he continues. “This drug, known as Pyrazinamide (PZA), shortens the treatment time for TB patients from up to a year to several months. Zimhony and his colleagues have found evidence that PZA and similar molecules can inhibit FAS-I. Now that the FAS-I structure is known, this mechanism can be tested. Other molecules can also be tested, and any potential drug molecules can then be further improved. Ideally, these would have an enhanced synergistic effect with antibiotics and expose the internal parts of the tuberculous bacilli to the immune system.
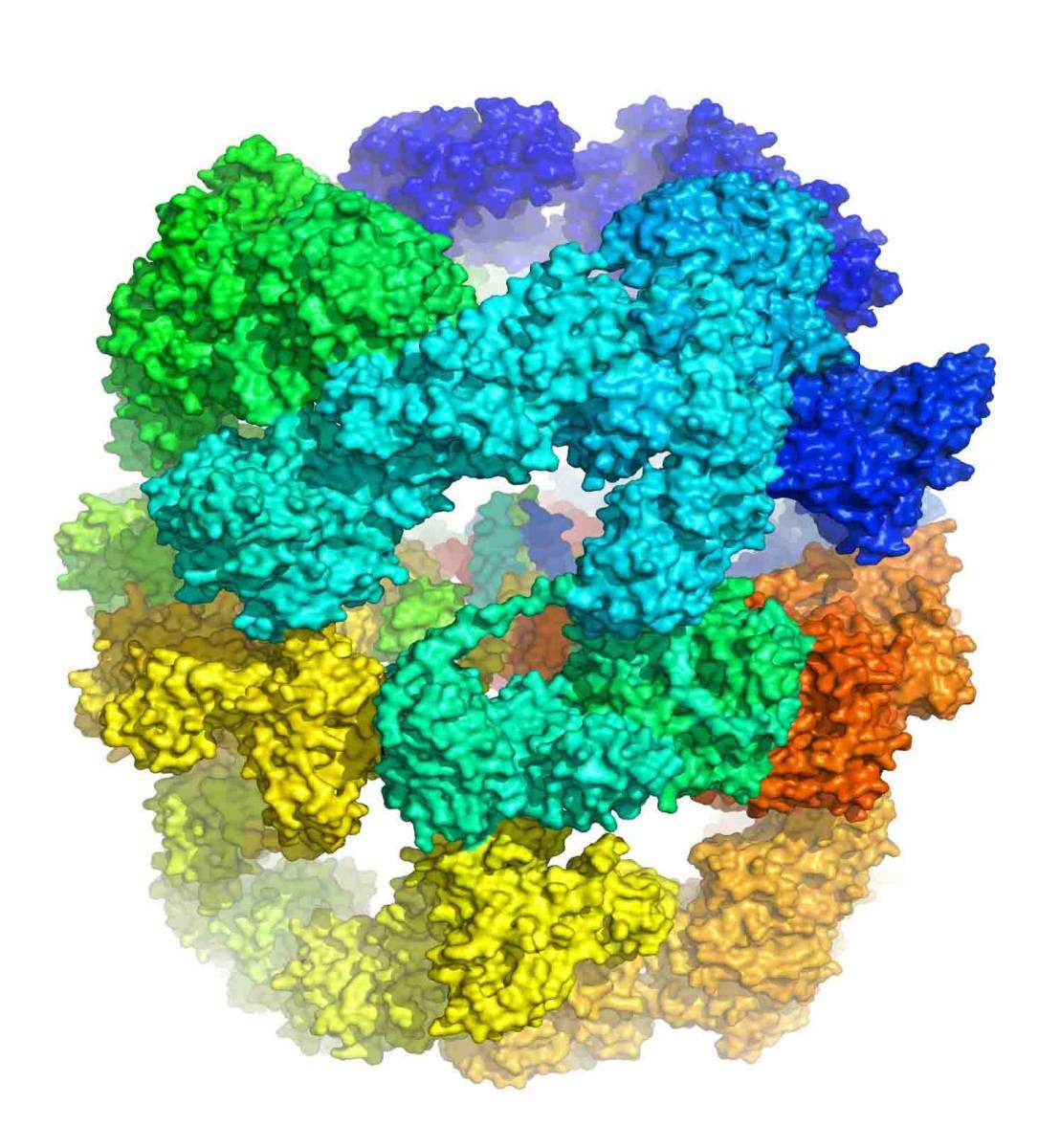
Surface representation of the ~2 MDa FAS-I from Mycobacterium tuberculosis. The six identical polypeptide chains that make the complete hexametric barrel-like complex are depicted with differ
“On a different note,” Diskin adds, “this project is a beautiful example of how one can achieve structural information for highly challenging targets, despite the experimental hurdles. It takes endless perseverance and a large team of dedicated and highly professional researchers.” Also participating in this research were Dr. Szilvia Baron and Gal Raviv of the Structural Biology Department; Drs. David Morgenstern and Yishai Levin of the de Botton Institute for Protein Profiling, a part of the Stephen and Nancy Grand IsraelNational Center for Personalized Medicine (G-INCPM); Dr. Sharon Wolf, Head of the EM Unit of Chemical Research Support; and Drs. Yoav Peleg, Shira Albeck, Jacob Grunwald and Orly Dym of the Structural Proteomics Unit.
The project was funded by the Kimmelman Center for Biomolecular Structure and Assembly.
Dr. Ron Diskin’s research is supported by the Moros Integrated Cancer Center; the Lester Crown Brain Research Fund; and the estate of Emile Mimran,
Prof Zippora Shakked’s research is supported by the Helen and Milton A. Kimmelman Center for Biomolecular Structure and Assembly.

Recent Comments