
Getting a fix on when and where mistakes occur in the cellular manufacture of proteins is so challenging that a researcher in the United States once declared he would take out to lunch for an entire year anyone who’d manage to pull it off. Prof. Yitzhak Pilpel, head of the Molecular Genetics Department at the Weizmann Institute of Science, couldn’t take him up on the offer because of the geographic distance, but when a recent study of his appeared in Molecular Cell, he emailed his American colleague to let him know: The formidable mission has been accomplished.
He and other scientists in Israel and Europe had not only succeeded in measuring the rate of mistakes occurring when proteins are made in the cell, they had also revealed that the DNA contains a “mistake manual” of sorts that dictates where these mistakes need to be avoided at all cost and where they may be tolerated or even welcome. Some mistakes, for example, might cause misfolded proteins to aggregate in a cell, as happens in Alzheimer’s disease, whereas others may give a cell an evolutionary advantage.
Errors in protein production can originate from spelling mistakes in the DNA itself, but much more common are mistakes occurring later in the production process, when the genetic message is copied and sent to the ribosome, the cell’s protein “factory.” Mapping out these late-stage mistakes has until now been nearly impossible, because all analyses yielded average numbers for all of the protein building blocks ‒ the amino acids ‒ in a cell, which may signal the presence of errors but not where on the protein those errors had occurred.

(l-r) Prof. Yitzhak Pilpel, Dr. Orna Dahan, Omer Asraf and Dr. Roni Rak
Pilpel, in collaboration with Prof. Tamar Geiger of Tel Aviv University and Prof. Ariel B. Lindner of Paris Descartes University, addressed this challenged by applying advanced algorithms to data obtained with a mass spectrometry method recently developed for studying individual substitutions of amino acids in a protein. The researchers then tested this approach on rapidly dividing yeast and bacterial cells. The result: They succeeded in detecting, quantifying and analyzing all the mistakes in cells’ proteins, down to the composition of individual amino acids. The research was conducted by Pilpel’s graduate student Ernest Mordret. The research team also included Dr. Orna Dahan, Omer Asraf and Avia Yehonadav of Weizmann’s Molecular Genetics Department; Dr. Georgina D. Barnabas of Tel Aviv University; and Prof. Jürgen Cox of the Max Planck Institute for Biochemistry.
It turned out that mistakes occur most commonly in the ribosome ‒ that is, in the final stage of protein production, known as “translation.” Wrong amino acids are inserted into a protein at this stage at the average rate of one in about 1,000 amino acids – that is, almost one mistake per protein. But the range is great: from one mistake in several dozen amino acids to one in about 10,000.
Mistakes are ‘allowed’ in positions where they are expected to be minimally detrimental
Perhaps the most surprising finding was that the distribution of these mistakes is far from random. They are much more common in proteins expressed in the cell in smaller rather than in larger amounts. This is probably because the latter, more abundant proteins would clutter up the cell much sooner if they contained errors, so that the cell would not have survived in the course of evolution. Moreover, within each protein, translation errors are much more common at positions that are less critical to the protein’s function and stability than at the more crucial ones – for example, those sites responsible for binding to other molecules.
“We found that mistakes are ‘allowed’ in positions where they are expected to be minimally detrimental, but not in the more sensitive positions,” Pilpel says.
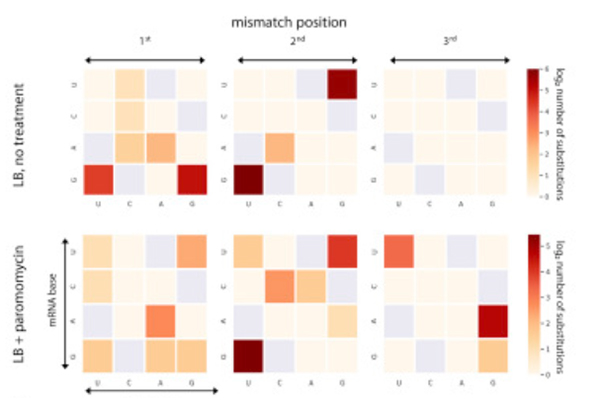
A mild dose of antibiotics (bottom) caused more mistakes to occur in translation, especially in certain positions. The darker the shade, the great the number of substitutions
But how does the ribosome know when it’s “allowed” to make mistakes? The study results suggest that the rate of mistakes at this stage is at least partially preprogrammed, and that such preprogramming might be achieved through the regulation of translation speed. In fact, the scientists found an inverse correlation between translation speed and accuracy. The faster the ribosome turned out proteins, the more errors it made. Conversely, it appeared to be programmed to slow down when accuracy was essential, as if it was taking its time in order to do things right. Translation speed, in turn, can be controlled via a genetic mechanism revealed in Pilpel’s earlier research. That mechanism has to do with the fact that the same amino acid can be encoded by different three-letter genetic sequences. Pilpel and his team had shown that some of the sequences – those that call for the use of abundantly available supporting molecules – enable the ribosome to pick up speed, whereas others call for the use of scarce molecules, forcing the ribosome to slow down until it finally gets the required molecule.
In the new study, the researchers also found that mistakes in translation can be triggered by external conditions. When they exposed dividing cells to an antibiotic, the rate of these mistakes increased.
These findings open up a new direction of research. Further studies could examine, for example, the role of translation mistakes in Alzheimer’s and other neurodegenerative diseases. Other studies could determine whether these mistakes slow down or accelerate cancerous growth, or whether they speed up the aging process.
The new findings may also help establish whether translation mistakes play a role in the evolution of species by creating diverse proteins that may help the organism adapt to changing conditions. “Mistakes in proteins generate diversity among genetically identical cells – a diversity that can prove beneficial in the course of evolution,” Pilpel says.

Recent Comments